Mechanism of Translation in Eukaryotic Cells
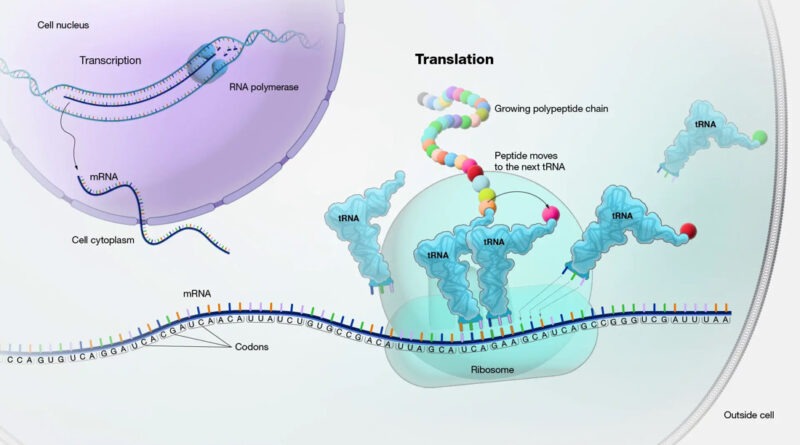
Translation is the biological process by which proteins are synthesized from mRNA templates, playing a crucial role in the expression of genes within eukaryotic cells. This process is intricate and involves various cellular components working in concert to ensure accurate and efficient protein synthesis. Here, we explore the stages of translation, the key players involved, and the regulatory mechanisms that ensure its fidelity and efficiency.
Overview of Translation
Translation occurs in the cytoplasm of eukaryotic cells and can be divided into three main phases:
1. Initiation
2. Elongation
3. Termination
Each of these phases involves specific steps and components, including ribosomes, transfer RNA (tRNA), messenger RNA (mRNA), and various protein factors.
Initiation
Initiation is a complex and highly regulated phase that sets the stage for protein synthesis. It begins with the formation of the pre-initiation complex, which includes the small ribosomal subunit (40S), initiation factors (eIFs), and the initiator tRNA charged with methionine (Met-tRNAi).
1. Formation of the Pre-Initiation Complex: The small ribosomal subunit binds with eukaryotic initiation factors (eIF1, eIF1A, eIF3, and eIF5) to form the 43S pre-initiation complex. This complex then associates with Met-tRNAi bound to eIF2-GTP, forming the 43S pre-initiation complex.
2. mRNA Binding and Scanning: The mRNA is prepared for translation by the addition of a 5′ cap and a 3′ poly-A tail. The cap-binding complex (eIF4F, which includes eIF4E, eIF4G, and eIF4A) recognizes and binds to the 5′ cap of the mRNA. The 43S complex is recruited to the mRNA, forming the 48S complex, which scans the mRNA in the 5′ to 3′ direction for the start codon (AUG).
3. Start Codon Recognition and Ribosome Assembly: Once the start codon is recognized, eIF2-GTP is hydrolyzed to eIF2-GDP, causing a conformational change that allows the large ribosomal subunit (60S) to join the complex. This assembly forms the 80S ribosome, ready for the elongation phase.
Elongation
During elongation, amino acids are sequentially added to the growing polypeptide chain. This phase involves three main steps: codon recognition, peptide bond formation, and translocation.
1. Codon Recognition: The ribosome has three sites: A (aminoacyl), P (peptidyl), and E (exit). The A site receives an aminoacyl-tRNA whose anticodon matches the mRNA codon. This process is facilitated by elongation factors eEF1A and eEF1B.
2. Peptide Bond Formation: The ribosome’s peptidyl transferase activity, located in the large subunit, catalyzes the formation of a peptide bond between the amino acid in the A site and the growing polypeptide chain in the P site. The polypeptide is then transferred to the tRNA in the A site.
3. Translocation: The ribosome moves one codon down the mRNA, shifting the tRNA in the A site to the P site and the tRNA in the P site to the E site. This movement is facilitated by elongation factor eEF2 and requires GTP hydrolysis. The empty tRNA in the E site is then released from the ribosome.
Termination
Termination occurs when a stop codon (UAA, UAG, or UGA) enters the A site. These codons do not correspond to any aminoacyl-tRNA but are recognized by release factors (eRF1 and eRF3).
1. Release of the Polypeptide: eRF1 binds to the stop codon and, together with eRF3-GTP, catalyzes the release of the newly synthesized polypeptide from the tRNA in the P site.
2. Disassembly of the Ribosome: Following polypeptide release, the ribosomal subunits, mRNA, and tRNA dissociate. This disassembly is assisted by ribosome recycling factors and ATP-dependent processes.
Regulatory Mechanisms
Translation in eukaryotic cells is tightly regulated at multiple levels to ensure protein synthesis occurs accurately and in response to cellular needs.
1. Initiation Regulation: The availability and activity of initiation factors (eIFs) are tightly controlled. For example, eIF2 is regulated through phosphorylation by kinases, which respond to various stress signals.
2. mRNA Surveillance: Quality control mechanisms, such as nonsense-mediated decay (NMD), no-go decay (NGD), and non-stop decay (NSD), ensure defective mRNAs are detected and degraded, preventing the production of faulty proteins.
3. Translational Repression and Activation: Specific RNA-binding proteins and microRNAs can repress or activate translation by binding to mRNA, influencing ribosome recruitment and scanning.
Conclusion
Translation is a vital and highly orchestrated process in eukaryotic cells, ensuring that genetic information is accurately converted into functional proteins. Understanding its mechanisms provides insights into cellular function and regulation, and highlights potential targets for therapeutic intervention in diseases related to protein synthesis dysfunction.